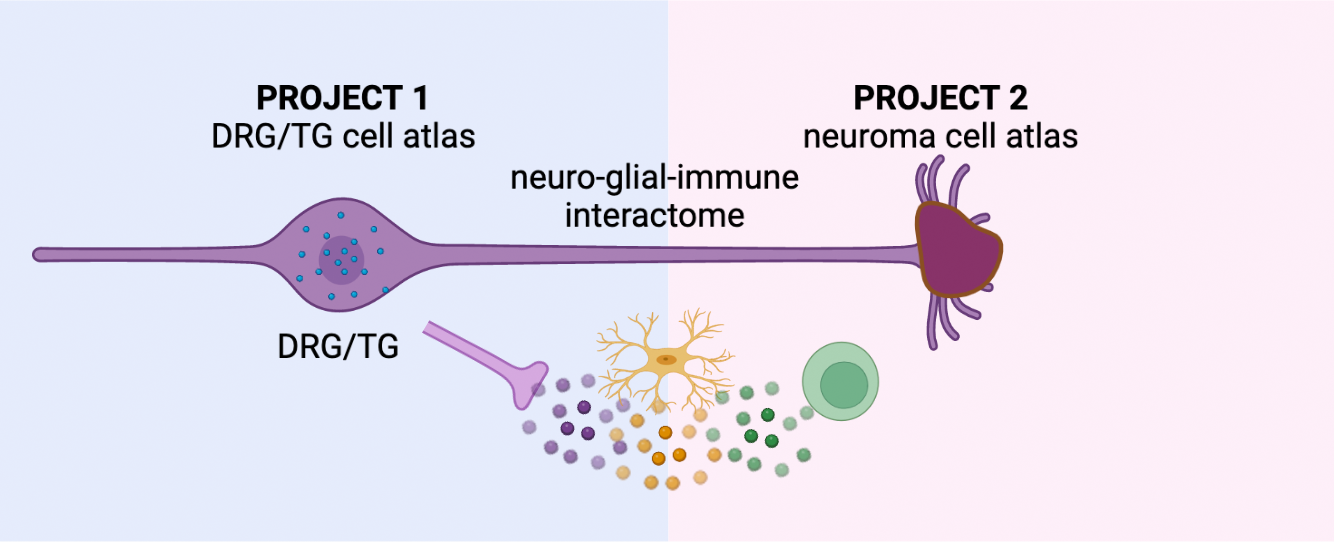
Project 1 – Multi-omic characterization of human nociceptors:
While we and others have recently generated the first single-cell profiles of human nociceptors, the sample sizes were insufficient to describe the full complement of molecules and epigenomic elements or the impact of natural genetic variation present in these cells. Project 1 will address this gap by identifying molecules in human nociceptors or nociceptor-associated non-neuronal cells that are central to pain signaling and could be targeted by novel pain therapeutics. Nociceptor epigenomic mapping will be used to interpret non-coding variants in pain GWAS. Together, these data could provide a basis for developing novel gene therapies and for understanding how genotypes and gene expression levels correlate with nociceptor physiology.
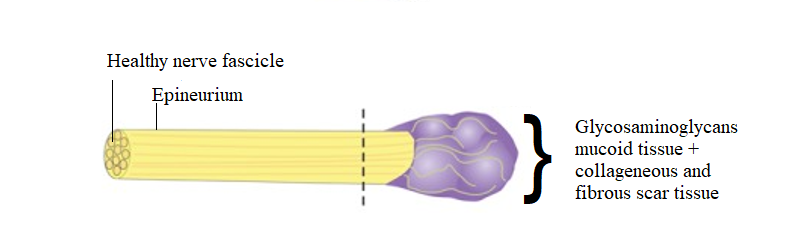
Project 2 – The cell types and states of painful human neuromas
Nerve injury is a major driver of chronic neuropathic pain and often leads to the development of neuromas, which occur when nerves are injured and are comprised of a mass of disorganized injured axons, Schwann cells, fibroblasts, blood vessels, and immune cells. In the case of limb amputation, multiple injured nerves branches will form neuromas. Not all neuromas are painful, and even in a single patient after a limb amputation, some neuromas may be more painful than others. Patients with painful neuromas often suffer from both acute and chronic pain. The responsible cell types, their transcriptional state, and the underlying molecular drivers of the pain in these neuromas are very poorly defined. Project 2 will address this gap by generating large single-cell datasets derived from patients undergoing excision of painful neuromas and targeted muscle reinnervation surgery or other intervention designed to reduce pain at the nerve ending. These data are particularly valuable because each neuroma is associated with a pain level in its sensory distribution, allowing for molecular correlates of pain to be assayed within the same individual. This genetically and self-controlled population of patients uniquely overcomes the profound heterogeneity of other “chronic pain” vs. “no pain” cohorts that could require exceedingly large sample sizes (e.g., tens of thousands) to separate signal from noise.